Special Issue: Water Narratives
24 December 2025
Published online 22 May 2023
High dimensional space representation of human diseases reveals relationships and predicts co-morbidities.
Andrey Rzhetsky
Enlarge image
Diseases are traditionally diagnosed and treated as independent phenomena. However, they often co-occur, with similar causes and overlapping symptoms, changing characteristics and outcomes of others.
Gengjie Jia of the Chinese Academy of Agricultural Sciences, and his colleagues, including researchers from King Abdullah University of Science and Technology in Saudi Arabia, applied computational methods to huge data sets to represent human diseases as inter-related nodes on a spectrum.
They used clinical records from 151 million Americans to map 547 disease categories on to a 20-dimensional space in which diseases with similar symptoms and causes lie close together. This was then mapped on to a three-dimensional metric space.
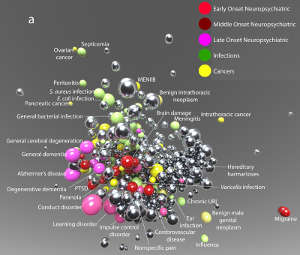
Here, diseases are classed as early-, middle- or late-onset neuropsychiatric conditions, infections, and cancers, with related diseases clustered together. Each disease is represented as an individual sphere, with the size of the sphere representing the prevalence of the disease.
The researchers also analysed genome-wide association data from 500,000 UK Biobank samples to identify 116 genetic associations with various health states, including 40 associations that had never been reported before. All the associations were linked to 108 loci, or chromosome positions.
By embedding each disease in a high-dimensional space, Jia and his colleagues discovered what they refer to as disease constellations, in which diseases are clustered with their closest neighbours.
They found, for example, that asthma lies in close proximity to allergic rhinitis, emphysema, chronic obstructive pulmonary disease, alveolar disease and pneumonia, all of which belong to the same constellation.
When the researchers mapped the genetic associations onto this space, they found that they could accurately predict co-morbidities, or which diseases will occur together, from other conditions within the same constellation.
“It would be fantastic to apply this clinically, but much work needs to be done before this is possible,” says senior author Andrey Rzhetsky, who specializes in bioinformatics research at the University of Chicago, USA. “The predictive power of genetic variation alone is limited, and environmental factors remain largely undiscovered.”
“Clinicians think about diseases as discrete entities, rather than continuous space,” he adds, “but one would have to generate a compelling proof that a new representation is a better view than the traditional one.”
doi:10.1038/nmiddleeast.2023.63
Jia, G. et al. The high-dimensional space of human diseases built from diagnosis records and mapped to genetic loci. Nat. Comput. Sci. https://doi.org/10.1038/s43588-023-00453-y (2023).
Stay connected: