Why I published a medical book for children
08 May 2025
Published online 11 May 2023
New genome sequencing effort captures much greater human diversity.

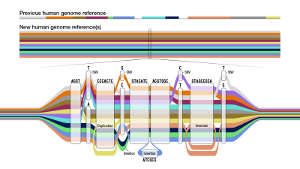
Darryl Leja, National Human Genome Research Institute, NIH
But that human genome represented the DNA of an incredibly small sample; most of it was from one individual and then only one chromosome in each pair, called the haplotype. While it was, for its time, the most complete human genome sequence available, there were significant gaps.
Now, an international consortium of researchers, including scientists from the Al Jalila Children’s Specialty Hospital, and the Mohammed Bin Rashid University of Medicine and Health Sciences, in Dubai, has published a draft pangenome made up of the complete human genome sequences from 47 individuals. More than half of those come from Africa, around one third from the Americas, 13% from Asia and 2% from Europe. They include people from ancestral backgrounds including Afro-Caribbean, Gambian, Peruvian, Punjabi, Han Chinese and Yoruba Nigerian.
The project started almost a decade ago as a conversation about how to build a reference human genome sequence that would be more representative of the diversity of humanity, says Benedict Paten, director of the Computational Genomics Lab at the University of California, Santa Cruz, and a co-lead author of the study.
“The reference is used as a lens to view pretty much all of genomics,” Paten says. “If you view everything through the lens of one set of sequences that don't necessarily represent the samples you're studying or the people that you're trying to sequence, then you bias yourself.”
But it would take multiple technological advances in both the sequencing and assembly of those multiple genomes to enable the project to achieve its first milestone in the publication of sequences from the first 47 of 350 individuals.
The first technological achievement is what co-author, Erik Garrison, calls “a quiet revolution in nanotechnology” that allows scientists to see single molecules of DNA. “The fact that you can have a single molecule means you can make a single read that is very, very long,” says Garrison, a computational genetics researcher at the University of Tennessee Health Science Center.
The sequencing technology that existed at the time of the first Human Genome Project enabled the reading of sequences the length of 100-150 base pairs – the paired nucleic acid molecules that are the fundamental units of DNA. But, technological advances now enable the sequencing of DNA strands that are tens, even hundreds of thousands of base pairs long.
Another technological advance is the field of computational genomics, which has enabled researchers to fit those genetic puzzle pieces together with far greater accuracy. “We think about methods to make computational genomics biology possible, in terms of how do we assemble genomes, how do we find variants from sequencing data, and how do we understand the differences that have occurred between genomes,” Paten says.
The research group also had to develop a novel way to present this wealth of genomic data and display the genetic variation seen across the many genomes. They achieved this with a genome graph in which variants in a particular sequence are shown expanded out into ‘bubble’ subgraphs.
“The genome graph is what we believe is, at this point, state-of-the-art for representing this, but it may not be the final answer,” says co-author Ting Wang, a geneticist and computational biologist at the McDonnell Genome Institute at the University of Washington, St Louis. “There are plenty of investigators trying to figure out if there are other ways to represent this data.” That also means developing tools to enable biomedical researchers to make the best use of this growing wealth of genetic data, Wang says.
For Olufunmilayo Olopade, oncologist and cancer geneticist at the University of Chicago’s Center for Clinical Cancer Genetics and Global Health, the pangenome is a “game changer” for her work on the variation in breast cancer genes in Black women and interactions with environmental factors.
“I'm truly delighted that we have this reference genome and that it is inclusive of a lot of diverse genomes,” says Olopade, who was not involved in the study. “This paper gives us more to think about as we design experiments looking at the heterogeneity of diseases.”
The pangenome project is now sequencing more genomes as the group works towards its goal of 350 end-to-end sequences of all chromosome pairs.
doi:10.1038/nmiddleeast.2023.49
Liao, W. et al. A draft human pangenome reference. Nature https://doi.org/10.1038/s41586-023-05896-x (2023).
Stay connected: